Northen Lab researchers Tami Swenson, Marc Van Goethem, and Trent Northen are authors on the paper “Learning representations of microbe-metabolite interactions” [1]. This research focuses on the integration of multiomics datasets and methods of finding relationships between microbial genes and metabolomes. A Matters Arising article by Quinn and Erb responded to the paper [2], and a following reply by the original authors was also published [3].
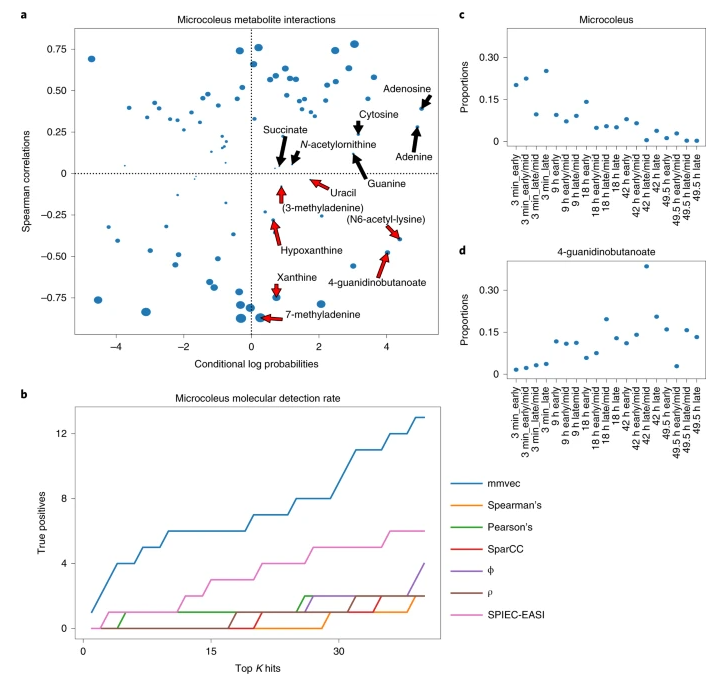
In their reply, the authors discuss the assertion by Quinn and Erb that correlation and proportionality can outperform the neural network mmvec in terms of identifying microbe-metabolite interactions. The method proposed as an alternative to mmvec involves the use of a centered log-ratio (CLR) transform before correlation analysis, which may not be applicable in situations where absolute abundance information is not available or data is sparse. Instead, mmvec utilizes co-occurrence probabilities instead of correlation, and outperformed the linear methods when the results of both were compared after being run on desert biocrust soils.
To learn more or view the entire discussion, check out the following links:
Learning representations of microbe–metabolite interactions
Examining microbe–metabolite correlations by linear methods
Reply to: Examining microbe–metabolite correlations by linear methods
References: