Northen Lab researchers Vlastimil Novak, Benjamin Bowen, Katherine Louie, and Trent Northen along with collaborators at University of California Berkeley, Joint BioEnergy Institute, LBNL, and Suranaree University of Technology co-authored the paper “Cynipid wasps systematically reprogram host metabolism and restructure cell walls in developing galls”. Galls are structures created by […]
Publications
Northen Lab scientists Peter Andeer, Kateryna Zhalnina, Lauren K. Jabusch, and Trent R. Northen recently co-authored an article published in Phytobiomes Journal titled, “Impact of Inoculation Practices on Microbiota Assembly and Community Stability in a Fabricated Ecosystem.” This research aimed to assess the challenge associated with different inoculation methods and […]
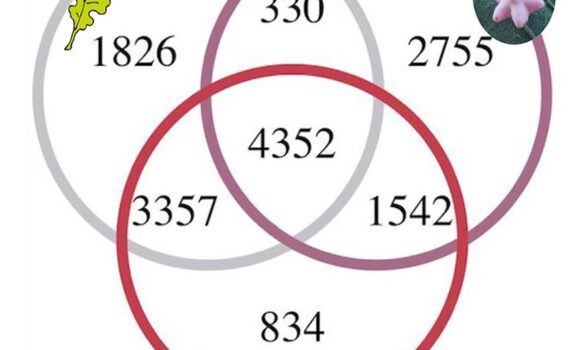
Northen Lab researchers Qing Zheng, Yuntao Hu, Suzanne M. Kosina, Marc W. Van Goethem, Susannah G. Tringe, Benjamin P. Bowen, and Trent R. Northen authored the publication “Conservation of beneficial microbes between the rhizosphere and the cyanosphere”. Biological soil crusts (biocrusts) are photoautotroph dominated communities on soil surfaces that provide […]
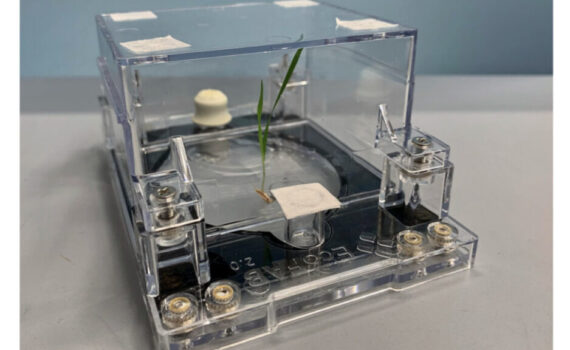
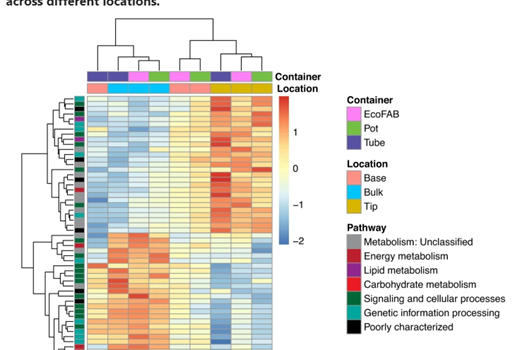
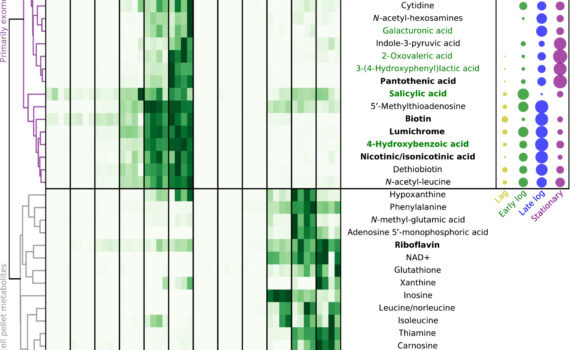
Northen Lab researchers Vlastimil Novak, Peter Andeer, Benjamin Bowen, Yezhang Ding, Kateryna Zhalnina, Connor Tomaka, Thomas Harwood, Michelle van Winden, Amber Golini, Suzanne Kosina, and Trent Northen, are authors on a new paper recently published in Science Advances in January 2024, titled “Reproducible growth of Brachypodium in EcoFAB 2.0 reveals […]
Northen Lab researchers Spencer Diamond, Peter Andeer, and Trent R. Northen and collaborators from the Lawrence Berkeley National Laboratory and the University of California, Berkeley, recently co-authored an International Society for Microbial Ecology (ISME) Communications paper: “Fine scale sampling reveals early differentiation of rhizosphere microbiome from bulk soil in young […]
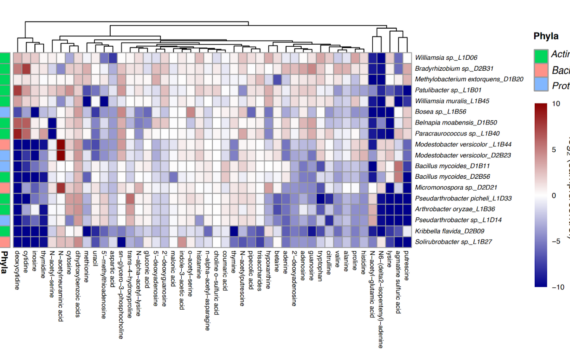
Northen Lab researchers Vanessa Brisson, Suzanne Kosina, and Trent Northen co-authored the publication “Dynamic Phaeodactylum tricornutum exometabolites shape surrounding bacterial communities”. While the role algal exometabolites play in regulating microbial community composition is not fully understood, this paper identifies algal exudates from the model diatom Phaeodactylum tricornutum and demonstrates their […]