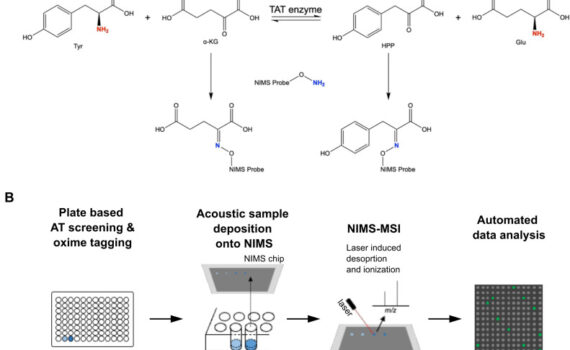
Northen Lab researchers Markus de Raad, Benjamin Bowen, Kai Deng, and Trent Northen recently authored a research article in the Journal of Biological Chemistry investigating a new high-throughput method for screening aminotransferase (AT) activity. Although aminotransferases play a central role in nitrogen metabolism for all species, the functionality of these […]
Publications
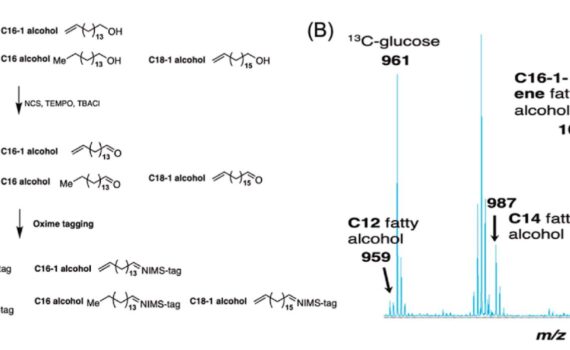
Northen Lab researchers Kai Deng, Markus de Raad, and Trent Northen along with collaborators at University of California Berkeley, Lawrence Livermore National Lab, and Sandia National Labs recently co-authored the publication “Rapid quantification of alcohol production in microorganisms based on nanostructure-initiator mass spectrometry (NIMS)”. NIMS is a surface-based MS technique, […]
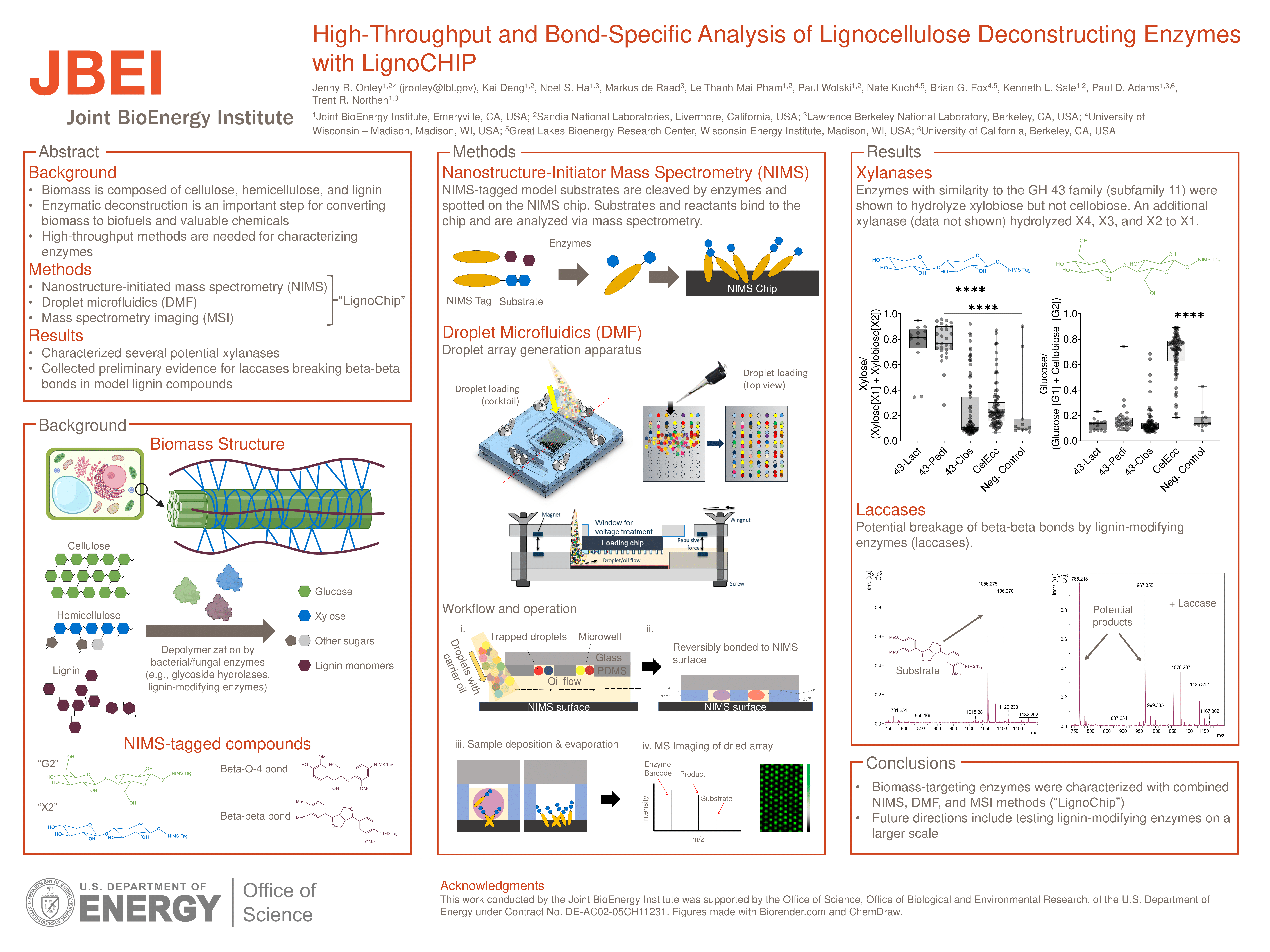
Northen Lab researcher Jenny Onley attended the 2022 ASM Microbe conference this past June, presenting the poster “High-Throughput and Bond-Specific Analysis of Lignocellulose Deconstructing Enzymes with LignoChip.” This technology was developed by a team of scientists including other lab members Kai Deng, Noel Ha, Markus de Raad, and Trent Northen. […]
The JGI metabolomics group and Northen Lab members (including Katherine Louie, Benjamin Bowen, Andrea Kuftin, and Trent Northen) have contributed to multiple recent papers. Some of the highlights include the following: Novel metabolic interactions and environmental conditions mediate the boreal peatmoss-cyanobacteria mutualism [1] Sphagnum (or peat moss) forms symbiotic relationships […]
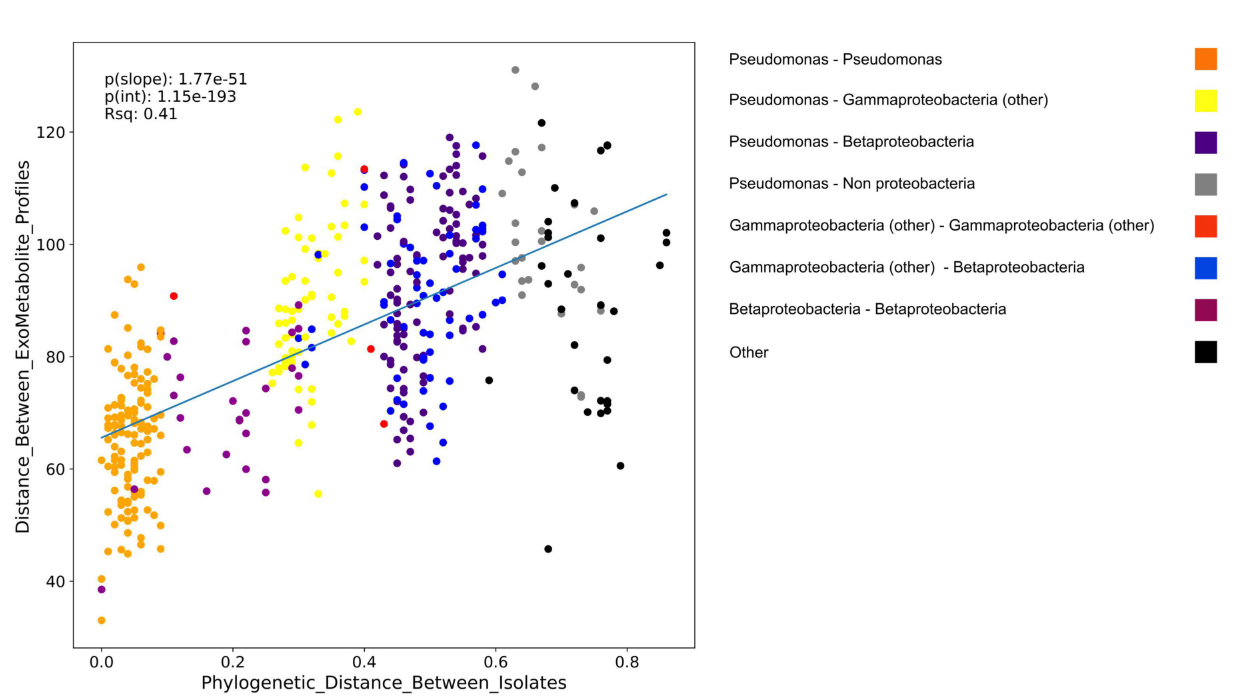
Northen Lab researchers Markus de Raad, Peter Andeer, Suzanne Kosina, Nicholas Saichek, Amber Golini, La Zhen Han, Ying Wang, Benjamin Bowen, and Trent Northen are authors on the paper “A Defined Medium for Cultivation and Exometabolite Profiling of Soil Bacteria.” Traditional media for microbial growth do not take into account […]
Northen Lab researchers Ying Wang, Tami Swenson, Anita Silver, Peter Andeer, Amber Golini, Suzanne Kosina, Benjamin Bowen, and Trent Northen are authors on the new paper “Substrate Utilization and Competitive Interactions Among Soil Bacteria Vary With Life-History Strategies.” This paper looks at genomic traits, exometabolite profiles, and interactions among soil […]