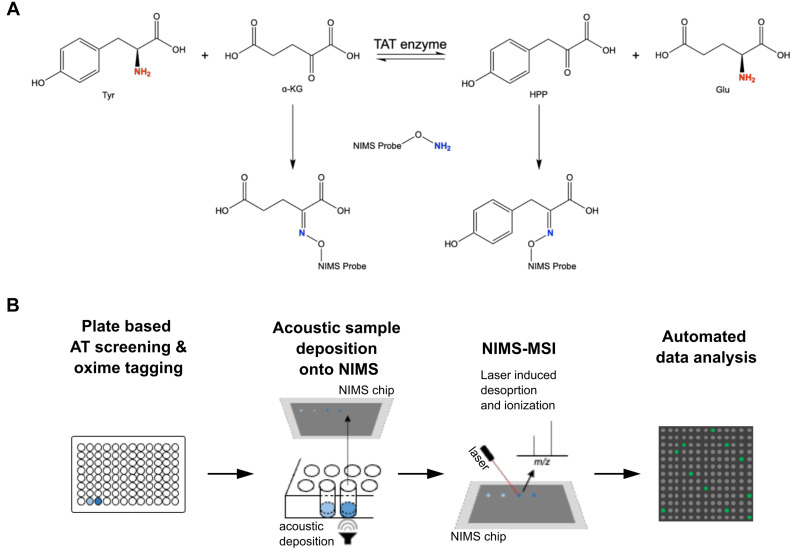
Northen Lab researchers Markus de Raad, Benjamin Bowen, Kai Deng, and Trent Northen recently authored a research article in the Journal of Biological Chemistry investigating a new high-throughput method for screening aminotransferase (AT) activity. Although aminotransferases play a central role in nitrogen metabolism for all species, the functionality of these enzymes is not well understood due to a lack of high-throughput assays that can effectively screen for substrate utilization. In this study, we present a new high-throughput technology for characterizing aminotransferase activity and specificity using mass spectrometry-based enzyme assay, that overcomes challenges with limited throughputs and specificities of standard methods.
Nanostructure-initiator mass spectrometry (NIMS) is used, in combination with a perfluorous alkoxyamine probe that forms an oxime linkage with ketones that are present in any keto acid products after transamination of amino donors. The tagged products are printed onto the NIMS surface using acoustic sample deposition, and analyzed using mass spectrometry imaging (MSI).Using this new method, it was discovered that the previously uncharacterized Arabidopsis thaliana tryptophan AT-related protein 1 enzyme can utilize 13 amino acid donors and three keto acid acceptors. This work demonstrates that this new high-throughput method enables screening of AT amino donor and keto acceptor specificity, thus providing a better understanding of the nitrogen metabolic network.
To learn more, read the full article here.
Reference:
de Raad, M., Koper, K., Deng, K., Bowen, B. P., Maeda, H. A., & Northen, T. R. (2023). Mass spectrometry imaging–based assays for aminotransferase activity reveal a broad substrate spectrum for a previously uncharacterized enzyme. Journal of Biological Chemistry, 299(3), 102939. https://doi.org/10.1016/j.jbc.2023.102939